How does Roche use machine learning to help understand pathology reports? This talk gives a high-level overview of their data pipeline and tech stack, including infrastructure they’ve built up to do OCR and NLP.
Speakers: David Talby (CTO @ Pacific AI), Vishakha Sharma (Principal Data Scientist @ Roche), Yogesh Pandit (Staff Software Engineer @ Roche)
The talk opened with a description of what Roche does and what kinds of problems they handle. This talk will focus on a pathology report, which has lots of jargon, lots of tables, and various key/value data. Manually curated reports look insane—

Looking at these, it was clear they’d need optical character recognition (OCR) to read what’s on the page and retain the document structure, and NLP (specialized for medical data and extensible for new content). The requirements for both were to support 10M pathology and radiation reports per year, comply with extensive privacy laws, integrate with AWS, and be low cost.
Healthcare data is quite complex and features lots of jargon—
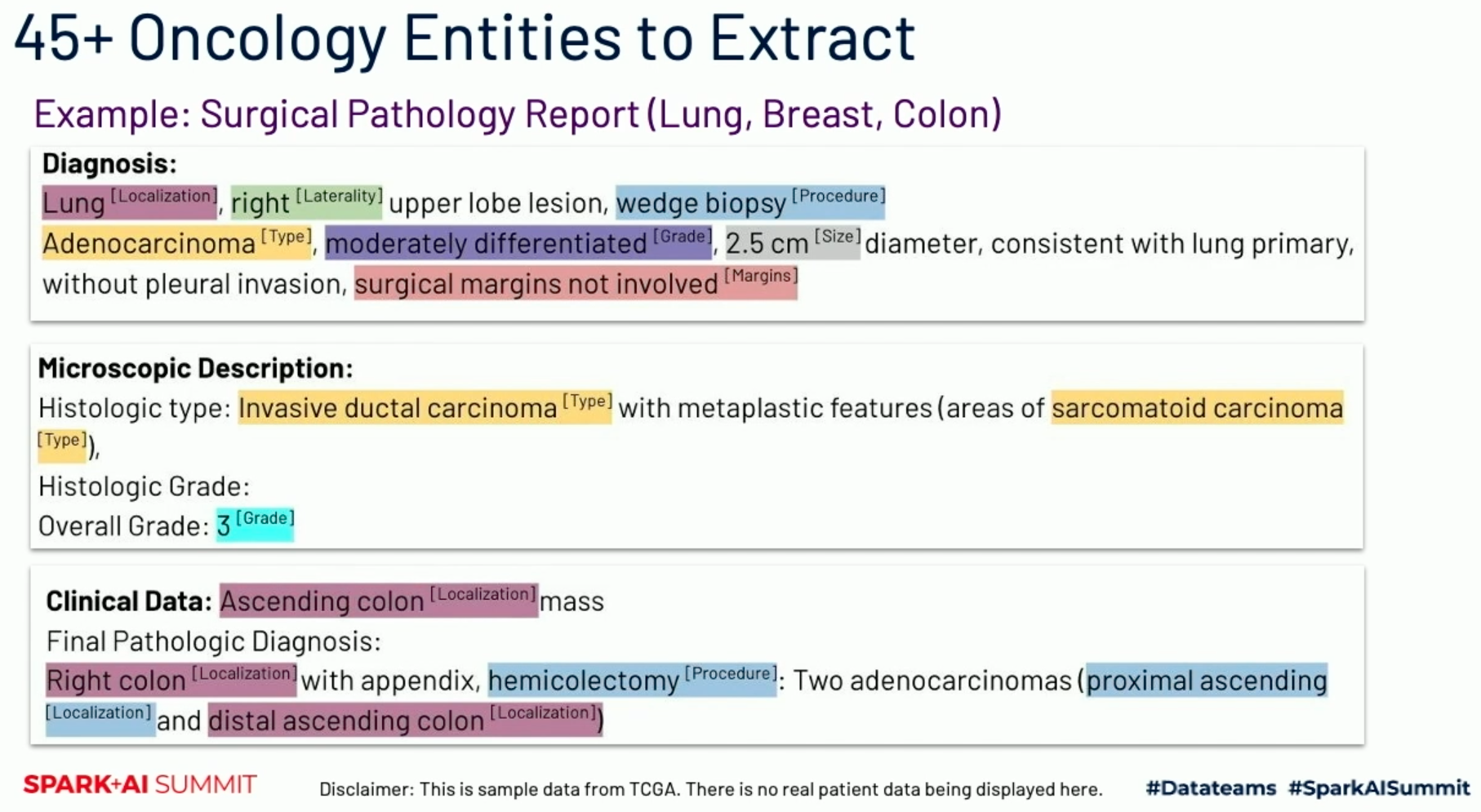
There are all kinds of things at play here, and these are interpretable only to doctors with years of training. There are types of issues (sarcomatoid carcinoma, adenocarcinoma), localizations (lung, kidney), orientations within the localizations, and more.
They used an OCR library for text extraction, evaluating it with word error rate, character error rate, and bag of words error rate. They used Named Entity Recognition (NER) using the pre-trained Bio-BERT to extract 45+ labels from pathology reports. Bio-BERT is included in Spark NLP.
Tools include Jenkins for orchestrating everything, including Jupyter notebooks-as-code through papermill (I love papermill!). They use MLflow to manage their model runs. A lot of things are more manual than you’d expect because of how much security they have to deal with.
The rest of the talk consisted of a discussion on Spark NLP, what it offers and why they chose to use it, some benchmarks, and other discussion about how they applied it. That was interesting, but I would have preferred more about their tech stack and architecture or about their modeling decisions.
Closing thoughts: This was a great talk! It struck the right balance of an interesting problem, reasonably novel techniques, engaging speakers, and high-quality slides (no scrolling through notebooks!).
